Comparing cortical data in low and high resolution scans
 |
The first step is to load the two surfaces we wish to compare
(File->Load surfaces, select the two .ply|.wrl files by
clicking on the first and ctrl-clicking on the second). wxRegSurf
decides which is the low resolution (LR, red) surface and which is the
high resolution (HR, green) surface by counting vertices. It is
important that the LR filename includes the string femur,
since there are some minor differences in the way wxRegSurf's
algorithms are tuned for different bone types. We now wish
to align these surfaces using a seven degree-of-freedom similarity
transformation (Registration->Similarity). The alternative is
to use a rigid registration, though the extra scale parameter allows
for scanner calibration errors and systematic segmentation
differences. wxRegSurf calculates a rough, initial alignment by
analysing the surfaces' shapes and topologies. If this fails, directly
manipulate the LR surface (Mouse->LR surface, left/middle mouse
button to rotate, right mouse button to translate, scrollwheel to
scale, single click to reflect) until the surfaces are roughly
aligned.
|
 |
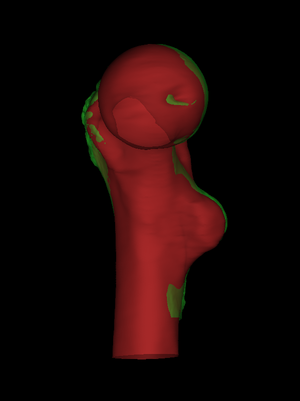
By default, wxRegSurf matches each vertex on the LR surface with the
closest vertex on the HR surface, as long as the distance between the
vertices is less than the Search range. To see these matches,
click Show matches. wxRegSurf's registration algorithms aim to
minimize the summed lengths of these matches. In this example, we see
a common complication with the LR surface extending below the HR
surface. If we registered with these matches, the LR surface would
rise too far up the HR surface, in a misguided attempt to minimize the
lengths of the matches at the bottom.
|
 |
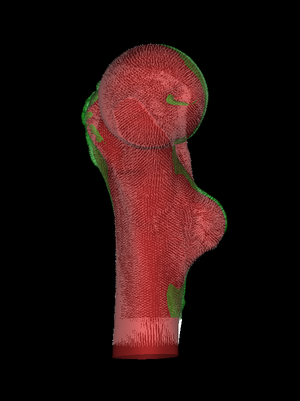
We could try to remove these unwanted matches by restricting the
Search range (and sometimes such adjustments are useful), but the
easiest way to proceed is to set Match rims to No. This
removes all matches between LR vertices and vertices on the open rim
of the HR surface. We now have an appropriate set of matches and are
ready to register.
|
 |
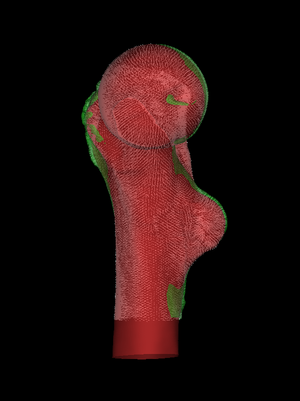
Next, we set the Iterations slider to something suitably high
and press Start. At each iteration, wxRegSurf finds the
transformation that minimizes the summed lengths of the matches and
then recomputes the matches. After a short while this process
converges to a stable alignment and we can either interrupt it
(Stop) or wait until the iteration count is
reached. File->Save registration saves the alignment in a
.reg file.
|
 |
The next stage of the analysis is to load the cortical data for the LR
surface (File->Load surface data, choose between the sw
thickness file, sw density file and surface data
file filters, then select the appropriate .bin
file). wxRegSurf will quietly load any corresponding error file it
finds in the same directory: these contain precision estimates at each
vertex and are used to weight any data smoothing we might perform. The
filenames for the cortical data must terminate in either
_thickness.bin (Stradview thickness), _density.bin
(Stradview density) or _dat.bin (general surface data).
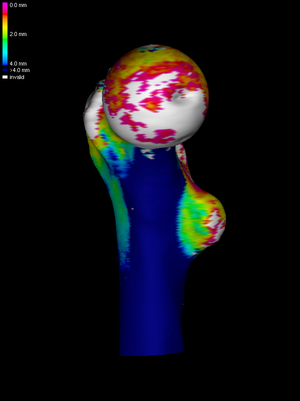
To see the data we have loaded, we select Surface
shading->Surface data. In this example, we see a rather poor estimate of
cortical thickness obtained by simple thresholding. The white areas
are where the CT data did not exceed the threshold.
|
 |
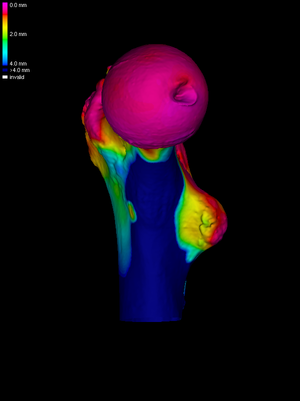
Next, we load the cortical data for the HR surface (File->Load
surface data again). To visualize this data clearly, without
the obscuring clutter of the LR data, we uncheck the Show LR
checkbox. It is clear that this data was measured at a much higher
resolution: the CT scans were performed in a high resolution pQCT
system, as opposed to conventional clinical CT for the low resolution
scans.
|
 |
wxRegSurf compares each LR data value with its matched HR value
(Show matches). For such a comparison to be meaningful, it is
first necessary to smooth the HR data until it matches the spatial
resolution of the LR data. Every time we press the Smooth
button, wxRegSurf replaces each data value with a weighted average of
immediately neighbouring values, with the weights set according to the
precision estimates found in the error file. If there is no error
file, uniform weighting is used. wxRegSurf smoothes any data that is
currently displayed, so if we only wish to smooth the HR data, we must
ensure that the Show LR checkbox is unchecked.
How much smoothing is necessary? This depends on the resolutions of
the scans and the average edge length in the HR mesh. We need to
calculate (elsewhere, wxRegSurf cannot help with this) how many
smoothing cycles are required to blur the high resolution point spread
function such that it approximates the low resolution point spread
function.
|
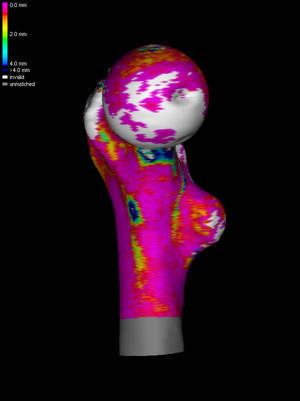 |
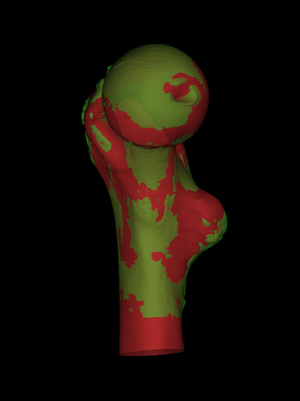
Finally, we can visualize the Signed data error or the
Absolute data error: the latter is shown on the
left. These errors are displayed on the LR surface, so we check
Show LR and uncheck Show HR. The results can be saved by
selecting File->Save LR-HR map. This writes a comma separated
variable (.csv) file with four columns. The first is the
index of each LR vertex, the second is the index of the matched HR
vertex (or "unmatched"), the third is the LR data which may be
"invalid", and the fourth is the (smoothed) HR data which may be
"unmatched" or "invalid". The .csv file is named
automatically, according to the two cortical data files that were
loaded and compared. It may be read into a spreadsheet or other
software for further analysis.
|
We might wish to repeat this process for tens or hundreds of
bones. wxRegSurf provides a mechanism for sequential processing using
batch (.run) files. An example batch file is shown below. We
could cut-and-paste these lines into a file called, for example,
MyBatchFile.run. A batch file can be run from the user
interface (File->Load and run batch file) or by supplying its
name as a command line argument when wxRegSurf is invoked.
/path/to/surface/and/data/files/including/trailing/slash/
# LR surface HR surface LR data HR data reflect smth LR smth HR range fussy n iter reg type save reg save LR data save map review rims save HR match
# -1=auto -=as->sm -1 = no 0 = no 0 = no surface colours
# 0 = yes 1-6 = angle 1 = yes
# >0 = smth - = LR only 2 = to rims
# register
lr_femur.wrl hr_femur.wrl n/a n/a 0 0 0 999 0 200 1 1 -1 0 0 0 0 0
# review
lr_femur.wrl hr_femur.wrl n/a n/a 0 0 0 999 0 200 1 0 -1 0 1 0 0 0
# map
lr_femur.wrl hr_femur.wrl lr_femur_thickness.bin hr_femur_thickness.bin 0 0 20 999 0 0 1 0 -1 1 0 0 0 0
A batch file may include any number of blank or comment lines. The
first non-comment line contains the full path to the working
directory, where wxRegSurf will look for .ply|.wrl and
.bin files. Each subsequent line contains a set of
instructions for wxRegSurf to load files, process them and then save
the results. Draft batch files can be produced using wxRegSurf's automatic batch file writer, then edited
manually as required.
In the example above, the first line loads two surface files (but
no thickness or density files) and registers them with a similarity
transformation (reg type is 0 for rigid, 1 for similarity, 2 for
similarity+FFD and so on, mirroring the options in the
Registration choice control). The search range is effectively
unconstrained (999) but rim matches are excluded (this is indicated by
the 0 in the penultimate column). 200 registration iterations are
performed before the results are saved in a .reg file. We
could fill the batch file with hundreds of similar lines, registering
different pairs of bones, and leave wxRegSurf to run unattended.
Following registration, we must check that all bones were aligned
satisfactorily. This is what the second line does in the example
above. It performs 200 iterations in "review" mode, whereby the
surfaces are loaded and spun around for the user to inspect. Lower
numbers of iterations reduce the inspection time, higher numbers
increase it. Alternatively, we could set the iteration count to zero,
in which case wxRegSurf instead saves a snapshot of the aligned bones
(.png) for offline inspection. Changing the review
flag from 1 to some other number saves a different view (1=front,
2=left, 3=back, 4=right, 5=top, 6=bottom). Any misregistered surfaces
must be corrected manually, with the updated results saved to the
.reg file.
The final line of the above example loads the two sets of cortical
data, performs 20 smoothing cycles on the HR data, applies the
similarity transformation from the .reg file, matches the LR
and HR vertices over an effectively unconstrained search range but
excluding rims, and then saves the LR-HR map in a .csv file.








