Sliding semilandmark registration
This tutorial describes details of an alternative registration
method for cortical bone mapping. It assumes that the reader is
already familiar with the general wxRegSurf workflow. If you have not
done so already, you should first read the femur tutorial.
When homologous point and curve landmarks can be identified on the
surfaces, you might prefer to use the sliding semilandmark method for
registering the surfaces. The sliding semilandmark algorithm is
commonly used for geometric morphometrics, and guarantees alignment of
the homologous features, while minimizing the bending energy of the
thin plate spline that warps the LR landmarks onto the HR landmarks.
 |
The first step is to manually identify landmarks on each of the
surfaces. wxRegSurf understands two types of manually localized
landmarks: points and curves. It is best to locate the landmarks using
some other software package that allows visualization of the
underlying data from which the surfaces were segmented. For example,
you might use Stradview to place landmarks with reference to the
underlying CT data. Stradview then exports the surface landmarks in the
corresponding .ply files. Alternatively, wxRegSurf has a
facility to place automatic landmarks on the surface of the proximal
femur. When a surface's filename includes the strings proximal and femur,
and the file does not already contain landmark definitions, wxRegSurf
will attempt to locate the centre of the fovea capitis, the
posterosuperior point on the intertrochanteric crest, the deepest
point of the trochanteric fossa, the head-neck boundary, the lesser
trochanter boundary, and the circumference of the femoral shaft at the
level of the lesser trochanter. In extremis, wxRegSurf also offers a
rudimentary landmark editor. Clicking on a surface while holding down
the shift key defines a point landmark. For open curves, press
o, then shift-click on points along the curve, then press
o again. For closed curves, press c instead of
o. The example on the left shows several points (large spheres)
and open curves (dashes) defined on an individual surface.
|
 |
Homologous landmarks, with matching names, must also be defined on the
canonical (LR) surface. In addition, a set of surface semilandmarks
(small spheres, left) is required to ensure that the LR surface stays
close to the HR surface everywhere. The surface semilandmarks should
be a suitable subset of the LR vertices. There is currently no
facility for generating them in wxRegSurf or Stradview, though the
femur and L1 vertebra canonical meshes, supplied with wxRegSurf, do
include suitable sets of semilandmarks. Surface semilandmarks do not
need to be defined on each individual, only on the canonical (LR)
mesh.
|
Landmarks are stored in .ply|.wrl files as comments. The
following .ply example should be self-explanatory:
comment LANDMARK "Radial extent of radial epicondyle" 0.13939 5.67478 57.00443
comment OPEN_LANDCURVE "Radial palmar condyle" 7
comment 4.28018 14.76402 56.43016
comment 3.12137 14.12788 55.24648
comment 2.30985 13.25270 54.62091
comment 1.42572 12.51070 54.90279
comment 0.76691 11.86253 55.92390
comment 0.70025 11.06597 57.07935
comment 0.79940 11.06698 58.44813
comment SEMILANDMARK 13.75 10.8253 4.70761
The syntax is the same for .wrl files, except that comment
lines start with <\tt># instead of comment. Note that
the stored landmark coordinates do not have to lie precisely on the
surface, since wxRegSurf projects them onto the surface when the
.ply|.wrl file is read in. Note also that landmark locations
may be specified using a single, integer vertex index, instead of the
(x,y,z) coordinates.
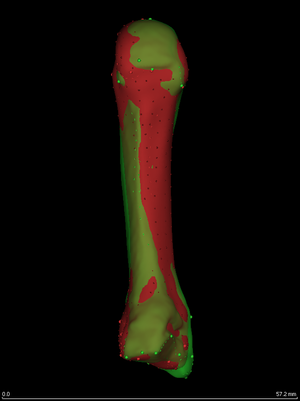 |
Here are the individual and canonical surfaces (third metacarpals),
aligned using wxRegSurf's Registration->Similarity option. Thus
far, the landmarks have not been used in any way. The surfaces have
just been brought into reasonably close proximity. But this is a good
starting point for the next stage. Selecting
Registration->Similarity+TPS, and pressing Start,
projects the surface semilandmarks from the LR surface onto the HR
surface, and the curve landmarks from the LR curves onto the
corresponding HR curves. Landmarks are then slid on the HR surface, so
as to minimize the bending energy of the thin plate spline that warps
the LR landmarks onto the HR landmarks. Surface semilandmarks can
slide in the surface tangent planes, while curve landmarks can slide
along tangents to the curves. Point landmarks do not slide. After
sliding, surface semilandmarks are projected back onto the HR surface,
and curve landmarks are projected back onto the corresponding HR
curve, ready for the next iteration.
|
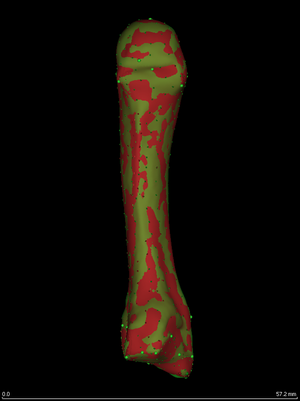 |
After twenty iterations, the sliding semilandmarks have converged to
stable positions. The homologous points and curves are perfectly
aligned. Note that curve landmarks are allowed to slide off the end of
open curves. If this behaviour is not appropriate, you should specify
the end points of the curves as point landmarks.
|
A suitable workflow for sliding semilandmark registration and
mapping, expressed as a batch
file, is provided below.
/path/to/surface/and/data/files/including/trailing/slash/
# LR surface HR surface LR data HR data reflect smth LR smth HR range fussy n iter reg type save reg save LR data save map review rims save HR match
# -1=auto -=as->sm -1 = no 0 = no 0 = no surface colours
# 0 = yes 1-6 = angle 1 = yes
# >0 = smth - = LR only 2 = to rims
# similarity registration
canonical_mc3_6756_lmarked.ply individual.ply n/a n/a 0 0 0 999 0 200 1 1 -1 0 0 1 0 0
# similarity+TPS registration
canonical_mc3_6756_lmarked.ply individual.ply n/a n/a 0 0 0 999 0 20 10 1 -1 0 0 1 0 0
# review
canonical_mc3_6756_lmarked.ply individual.ply n/a n/a 0 0 0 999 0 200 10 0 -1 0 1 1 0 0
# map
canonical_mc3_6756_lmarked.ply individual.ply n/a individual_thickness.bin 0 0 2 999 0 0 10 0 4 0 0 1 0 0




