Main > GMT_4YP_19_3
Dr Graham Treece, Department of Engineering
F-GMT11-1: Fast simulation of CT scanning in 3D
 | 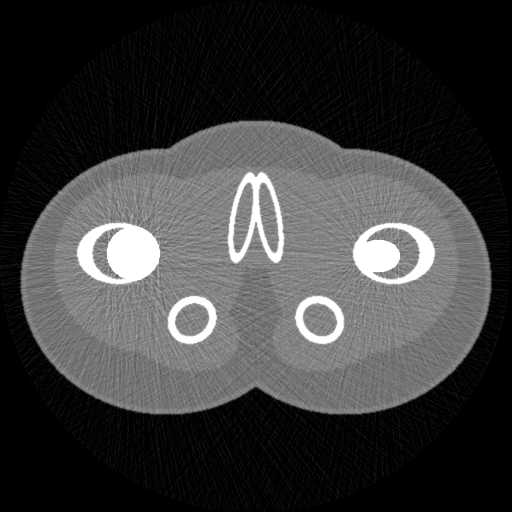 | 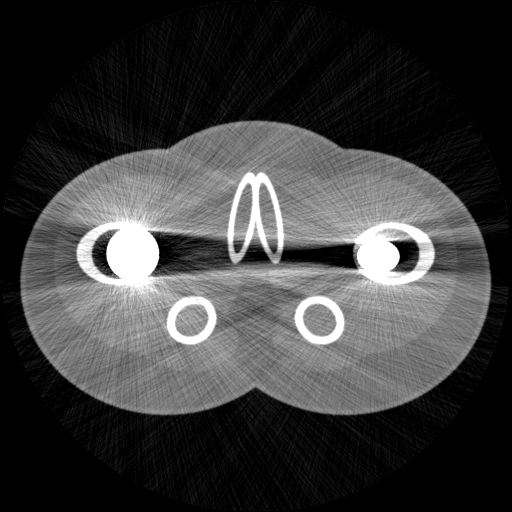 |
| This is a simulated 2D CT scan of a 'phantom' which is designed to look a bit like a scan of a pelvis. Though the geometry is made up, the data for the materials (bone, muscle, etc) is from experimental measurements. | Whilst the simulation is simple, it is possible to mimic the type of poisson noise actually present in normal CT scans. | The simulation also models more complex effects due to scattering and non-linearity, despite being reasonably fast: this scan took a few seconds to simulate. |
Computed Tomography (CT, effectively a 3D X-ray) scanning is frequently used in medicine for disease diagnosis. Current research in this area includes very precise methods for assessing bone in CT data, and also techniques for removing imaging problems due to the presence of metal implants. These developments can be very hard to test, since there is no clinically viable 'gold standard' CT scan to compare the results with. Hence CT simulation (mimicking a CT scan of a known, computer-defined, object, using software) is a vital tool in testing how well such techniques work. CT simulation is also used to help plan radiotherapy treatments for cancer.
'Proper' CT simulation software models the individual interactions of each photon in the X-ray beam with each tissue in the body: such systems understandably take a very long time to run, even when running on highly parallelised fast graphics processing units. However, for many purposes, a faster simulation technique is possible, based on the statistical properties of the X-ray beam as a whole, rather than individual photons, and presuming straight-line interactions with the tissue. An example of this sort of simulation is in the figure above, which runs either using python or Matlab, in a few seconds on a laptop.
However, this simulation is only in 2D: real clinical CT machines acquire 3D data, and they do it in different ways, for instance using cone-beam scanning, or helical CT. These techniques introduce specific (and different) problems into the reconstructed data. Is it possible to produce a 3D simulator which can mimic some of these, whilst still remaining fast enough to be practically useable?
This is an algorithmic development / computational geometry / software project, so experience of writing software is essential, though the development could be done using python, Matlab, or another programming environment.
Click here for other medical imaging projects offered by Graham Treece.