Dr Graham Treece, Department of Engineering
Sequential Freehand 3-D Ultrasound
Background
In the last two decades, a great many researchers have produced systems which allow the construction and visualisation of three dimensional (3-D) images from medical ultrasound data. Although many clinical applications for this emerging technology have been suggested, it is clear that sufficiently compelling ones are still to be found. One of the most promising areas where 3-D ultrasound can provide a real benefit is in the accurate measurement of volume. Volume measurement is important in several anatomical areas, for instance the heart, the foetus, placenta, kidney, prostate, bladder and the eye. Measurements have traditionally been made with 2-D ultrasound, but it is generally accepted that 3-D ultrasound can provide much greater accuracy. Freehand 3-D ultrasound, unlike other 3-D ultrasound techniques, allows the clinician unrestricted movement of the ultrasound probe. The ultrasound images (B-scans) are digitised with a video card and stored in a computer. In addition, the position and orientation of the probe is measured and recorded with each B-scan. One of the disadvantages of freehand scanning, compared to the other techniques, is that the recorded B-scans are not parallel, and may intersect each other. This makes processing of the data quite complex, hence most systems firstly interpolate this data to a regular 3-D array. However, this interpolation can take considerable time, and generate unwanted artifacts. It would be an advantage to be able to measure organ volume directly from cross-sections defined in the original freehand 3-D ultrasound B-scans. These B-scans do not contain processing artifacts, and hence the clinician has a better chance of accurately outlining the cross-sections of the organ (segmenting). In addition, estimating volumes and surfaces directly from these cross-sections should reduce the amount of processing time required, and allow measurements to be performed during, rather than after, the examination. It would also be an advantage to calculate accurate volumes from a smaller number of cross-sections, since manual segmentation, which is still the only universally reliable method for ultrasound data, is the most time consuming of the processes involved.Research
Three novel techniques are presented. A much more detailed description of cubic planimetry and maximal disc guided shape based interpolation can be found in a technical report, and of regularised marching tetrahedra in another technical report. Both these papers have also been published in journals.Cubic planimetry
 | 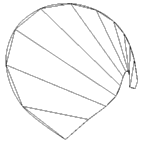 |  |
| (a) Cross-sections | (b) 2-D | (c) Volume |
Figure 1. Cubic planimetry: sphere. Once the cross-sections have been segmented as in (a), they are reduced to a 2-D graph (b) from which the volume can be calculated (c).
Maximal disc guided shape based interpolation
 |  | 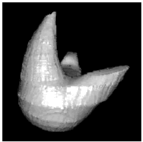 |
| (a) Cross-sections | (b) Discs | (c) Surface |
Figure 2. Maximal disc guided shape based interpolation: simulated object. A disc based representation of the original cross-sections in (a) is first created (b). This is used, in conjunction with a distance transform of each cross-section, to interpolate the object. (c) Surface points and normals can be extracted from the interpolated data for a simple rendering of the object.
Regularised marching tetrahedra
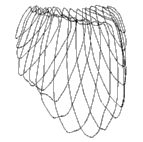 |  | 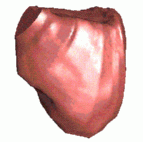 |
| (a) Cross-sections | (b) Triangle mesh | (c) Surface |
Figure 3. Regularised marching tetrahedra: human bladder. Rather than interpolating the cross-sections in (a) to an array, they can be gradually interpolated to a tetrahedral lattice. (b) The iso-surface is extracted and triangulated during this process. (c) The regular aspect ratios of the triangles leads to a high quality image when interpolated shading is used to render the surface.
Summary
These three algorithms provide the clinician with high quality interactive surface renderings and an accurate volume estimation, from segmented object cross-sections. The processing is completed fast enough to be performed during the examination, rather than afterwards. Currently, the segmentation is performed using a computer assisted manual method, which typically takes 30secs per object cross-section. All of the above techniques have been integrated into Stradwin, a real time freehand 3-D ultrasound acquisition and visualisation system, and IsoSurf, which is a more generic package for parallel data.- Graham Treece
- Introduction
- Teaching
- Research
- Publications
- Software
- Videos
- Personal
- Medical Imaging Group
- Overview
- Members
- Projects
- Research Opportunities
- Free Software
- Machine Intelligence Homepage